 Restriction endonucleases cleave DNA at specific sites
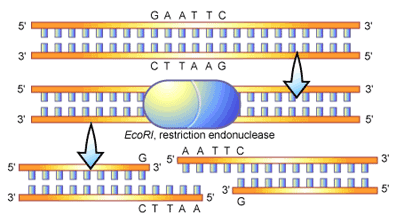
Restriction endonuclease Eco RI, cleaves the sequence 5'GAATTC3', was initially isolated from bacteria (Escherichia coli). This restriction endonuclease will cleave DNA isolated from any cell, prokaryotic or eukaryotic.
Restriction endonucleases cleave DNA at specific sites
Restriction endonuclease Eco RI, cleaves the sequence 5'GAATTC3', was initially isolated from bacteria (Escherichia coli). This restriction endonuclease will cleave DNA isolated from any cell, prokaryotic or eukaryotic. One of the first steps to manipulate and study discrete DNA sequences is the ability to cleave large molecules of DNA into smaller defined fragments. This can be accomplished by the use of restriction endonucleases, which are enzymes that can cleave (or digest) double – stranded DNA in a site – specific fashion.
These enzymes are found naturally in a variety of different bacteria, and are used by the bacteria to degrade foreign DNA; their own DNA is modified to be protected from degradation. Generally, restriction endonucleases recognize a short nucleotide sequence (4 – 8 bp) and cleave the DNA in a precise fashion. Cleavage breaks the phosphodiester bond in both of the two strands of double – stranded DNA. Many restriction endonuclease recognition sites are palindromic, meaning the sequence is identical whether reading from the top or bottom strand (but always reading 5' to 37apos;).
For example , the restriction endonuclease Eco RI, which cleaves the sequence 5'GAATTC3', was initially isolated from bacteria ( Escherichia coli). This restriction endonuclease will cleave DNA isolated from any cell, prokaryotic or eukaryotic. There are dozens of different restriction endonucleases that recognize and cleave different sequences.